pUNO1-SARS2-M
-
Cat.code:
puno1-cov2-m
- Documents
SARS-CoV-2 Matrix/Membrane coding sequence
GENE DESCRIPTION
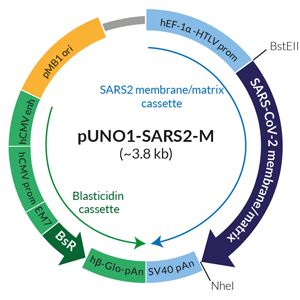
Schematic of SARS-CoV-2 membrane/matrix expression vector
Matrix/Membrane (M) is a type III transmembrane glycoprotein that is the most abundant among SARS-CoV-2 structural proteins. M, Spike(S), and Envelope (E) proteins, constitute coronaviruses’ interface to the external environment [1, 2]. The dimerization of M proteins and their interaction with other structural proteins (notably E) is responsible for the virus sphere shape, and the formation of a matrix-like layer underneath the viral membrane [1, 2]. M is also described as the primary driver of coronaviruses budding process [2].
pUNO1-SARS2-M contains the wild-type coding sequence of SARS-CoV-2 Matrix/Membrane. (See Details and Specifications for more information).
 InvivoGen also offers:
InvivoGen also offers:
• SARS-CoV-2-Cellular Receptor Genes
• SARS-CoV-2 S, S1, RBD, E, N Genes
PLASMID DESCRIPTION
pUNO1-SARS2-M features a potent mammalian expression cassette comprised of the ubiquitous human EF1α-HTLV composite promoter and the SV40 polyadenylation (pAn) signal. The ORF is flanked by unique restriction sites BstEII and NheI) to facilitate its subcloning. The plasmid is selectable with blasticidin in both E. coli and mammalian cells. It can be used for transient or stable transfection. It contains no tag.
QUALITY CONTROL
- Fully sequenced ORF
- Predominant supercoiled conformation
![]() Learn more about SARS-CoV-2 infection cycle, immune responses, and potential therapeutics.
Learn more about SARS-CoV-2 infection cycle, immune responses, and potential therapeutics.
References
1. AJ Alsaadi E. & Jones I.M., 2019. Membrane binding proteins of coronaviruses. Future Virology. 14(4):275-286.
2. Neuman B.W. et al., 2011. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174:11-22.
Specifications
- Strain: Wuhan-Hu-1 isolate
- Genbank: NC_045512.2
- ORF size from ATG to Stop codon: 669 bp
- Native (wild-type) sequence
-
Subcloning restriction sites in pUNO1: BstEII (in 5’) and NheI (in 3’)
- BstEII generates cohesive ends. BstEII has no known compatible restriction enzyme.
- NheI generates cohesive ends compatible with AvrII, SpeI, and XbaI restriction sites -
Sequencing primers:
- Forward HTLV 5’UTR: TGCTTGCTCAACTCTACGTC
- Reverse SV40 pAn: AACTTGTTTATTGCAGCTT
Contents
- 20 μg of lyophilized DNA
- 2 x 1 ml blasticidin at 10 mg/ml
![]() The product is shipped at room temperature.
The product is shipped at room temperature.
![]() Lyophilized DNA should be stored at -20 ̊C.
Lyophilized DNA should be stored at -20 ̊C.
![]() Resuspended DNA should be stored at -20 ̊C and is stable up to 1 year.
Resuspended DNA should be stored at -20 ̊C and is stable up to 1 year.
![]() Blasticidin is a harmful compound. Refer to the safety data sheet for handling instructions.
Blasticidin is a harmful compound. Refer to the safety data sheet for handling instructions.
Store blasticidin at 4°C or -20°C for up to two years. The product is stable for 2 weeks at 37°C.
Avoid repeated freeze-thaw cycles.
Details
The current knowledge about the M protein comes from previous studies on β-coronaviruses other than SARS-CoV-2, including SARS-CoV and mouse hepatitis virus (MHV).
The M protein is a type III transmembrane glycoprotein of approximately 230 amino acids (a.a) and is comprised of three regions: an N-terminal ectodomain, three transmembrane domains, and a C-terminal endodomain [1].
M proteins are present as an MLONG and MCOMPACT population and are functionally dimeric in viral particles [2]. It has been suggested that the interaction between MLONG dimers by their endodomains forms a matrix-like layer underneath the viral membrane [2].
M is the primary driver of coronaviruses budding process [2]. MLONG and MCOMPACT are present in the endoplasmic reticulum and are transported to the budding site where they interact with other viral membrane proteins. Increasing density of MLONG through the conversion of MCOMPACT allows the bending of the virus membrane to form a sphere around the nucleocapsid and viral RNA [2].
During the assembly of new virions, M interacts with itself and the other structural proteins, Envelope (E), Spike (S), and Nucleocapsid (N) [1, 2].
References
1. AJ Alsaadi E. & Jones I.M., 2019. Membrane binding proteins of coronaviruses. Future Virology. 14(4):275-286.
2. Neuman B.W. et al., 2011. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174:11-22.
DOCUMENTS
Documents
Technical Data Sheet
Plasmid Map and Sequence
Safety Data Sheet
Plasmid Sequence
Certificate of analysis
Need a CoA ?

