A549-ASC-GFP-NLRP1 Reporter Cells for COVID-19 Studies
| Product | Unit size | Cat. code | Docs. | Qty. | Price | |
|---|---|---|---|---|---|---|
|
A549-ASCoV2-NLRP1 Cells Human ACE2, TMPRSS2 & NLRP1 lung reporter cells |
Show product |
3-7 x 10e6 cells |
a549-ascov2-nlrp1
|
|
||
|
A549-ASCoV2-NLRP1 vial Additional cell vial |
Show product |
3-7 x 10e6 cells |
a549-ascov2-nlrp1-av
|
|||
|
A549-ASCoV2 Cells Human ACE2 & TMPRSS2 lung reporter cells |
Show product |
3-7 x 10e6 cells |
a549-ascov2
|
|
||
|
A549-ASCoV2 vial Additional cell vial |
Show product |
3-7 x 10e6 cells |
a549-ascov2-av
|
Notification: Reference #a549-ascov2-nlrp1-av can only be ordered together with reference #a549-ascov2-nlrp1.
Reference #a549-ascov2-av can only be ordered together with reference #a549-ascov2.
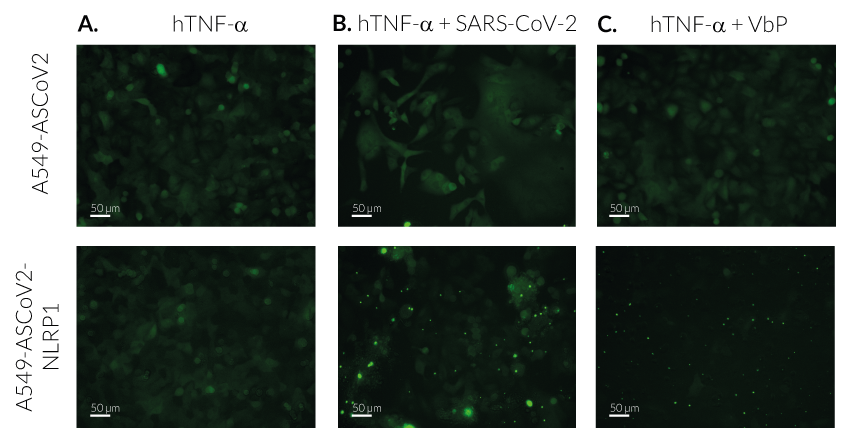
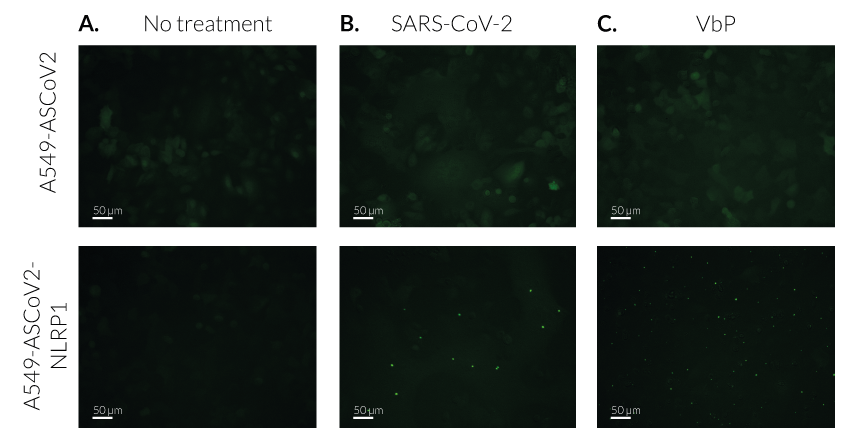
SARS-CoV-2-mediated ASC speck in A549-ASCoV2-NLRP1 cells
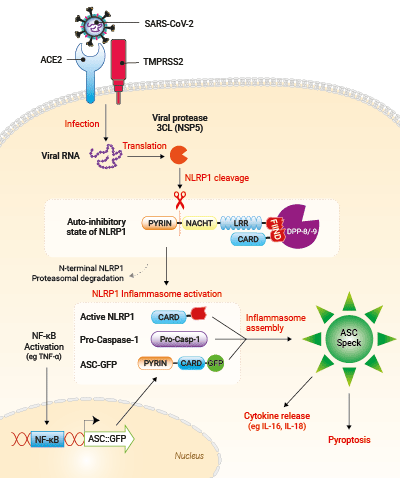
Upon SARS-CoV-2 infection, viral RNA is released in target cells and immediately translated to produce viral polyprotein including the 3CL protease (NSP5). In addition to process viral polyprotein, 3CL cuts NLRP1 in its N-terminal domain. Cleavage creates an unstable neo-N terminus that is rapidly degraded by proteasome pathway, hence releasing the C-terminal part of NLRP1. Free NLRP1 C-terminal domain recruits ASC::GFP and Caspase-1 leading to the NLRP1 inflammasome assembly. This triggers the formation of ASC-GFP specks, the release of cytokines (eg IL-16 and IL-18) and pyroptotic cell death.
Study of SARS-CoV-2-dependent NLRP1 inflammasome activation
InvivoGen offers a human A549 lung carcinoma-derived cell line specifically designed for assessing ASC-dependent NLRP1 inflammasome activation upon SARS-CoV-2 infection and its control cell line:
— A549-ASCoV2-NLRP1 cells
— A549-ASCoV2 (control) cells
- A549-ASCoV2 NLRP1 cells and their control cell line feature an NF-κB-inducible ASC::GFP reporter gene. This allows the monitoring of ASC-dependent inflammasome activation by visualizing ASC speck formation using fluorescence microscopy.
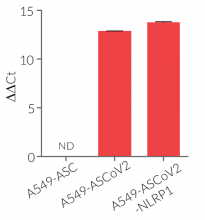
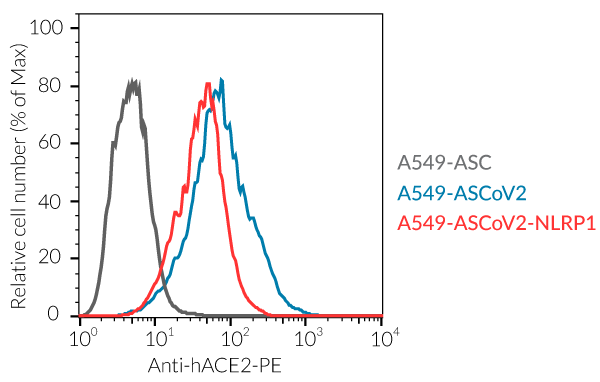
- A549-ASCoV2 NLRP1 cells and their control cell line overexpress the genes encoding the SARS-CoV-2 human receptors, ACE2 and TMPRSS2 [1], and are thus highly permissive to SARS-CoV-2 infection.
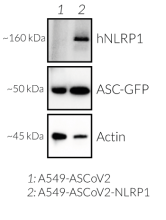
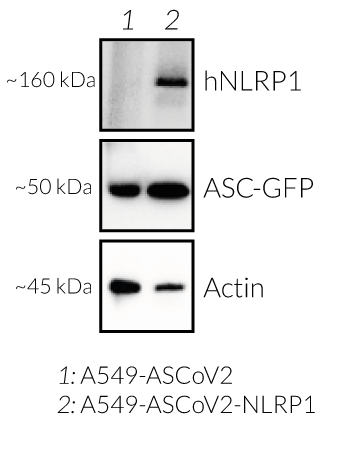
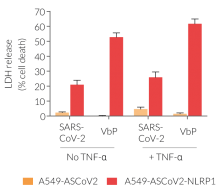
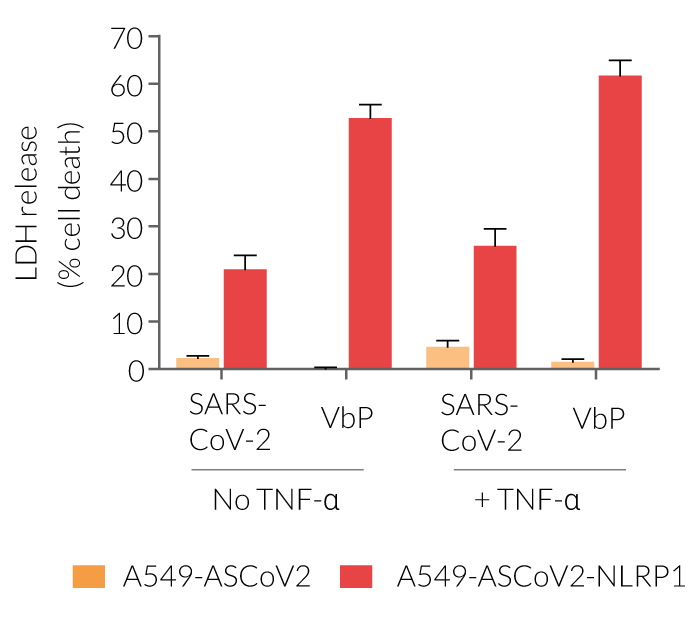
- A549-ASCoV2-NLRP1 cells have been additionally engineered to stably express the human NLRP1 inflammasome sensor. Its activation can be monitored in real-time by following the formation of fluorescent ASC specks and measuring pyroptotic cell death with the LDH assay.
The NLRP1 inflammasome has been shown to play an important role in COVID-19 pathophysiology. Indeed, the NLRP1 sensor is activated upon cleavage by the SARS-CoV-2 3CL protease NSP5 in lung epithelial cells [2].
Upon induction of ASC::GFP with the NF-κB inducer TNF-α and stimulation using SARS-CoV-2 infectious particles, A549-ASCoV2-NLRP1 cells display ASC specks, visible as fluorescent green dots and undergo pyroptosis, readily measurable by the LDH assay.
A549-ASCoV2 cells not only serve as a control cell line for A549-ASCoV2-NLRP1 cells but also offer the possibility to monitor ASC-dependent inflammasome activation of NLRP3 after its transgenic expression.
Key features:
- NF-κB-dependent expression of the ASC::GFP fusion protein
- Stable expression of human NLRP1 (isoform 1)
- Overexpression of human ACE2 and TMPRSS2
- Highly permissive to SARS-CoV-2 infection
Applications:
- Screening of NLRP1 inflammasome activating stimuli
- Real-time monitoring of ASC-dependent NLRP1 inflammasome assembly
- Rapid and visual screening of COVID-19 drugs targeting the NLRP1 pathway
![]() Read our review on NLRP1 & NLRP3 inflammasome sensors
Read our review on NLRP1 & NLRP3 inflammasome sensors
1. Hoffmann M. et al., 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:1.
2. Planès R. et al., 2022. Human NLRP1 is a sensor of pathogenic coronavirus 3CL proteases in lung epithelial cells. Mol Cell. S1097-2765(22).
Specifications
Growth medium: DMEM, 4.5 g/L glucose, 2 mM L-glutamine, 25 mM HEPES, 10% (v/v) heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin, 100 µg/ml Normocin™
Antibiotic resistance: Blasticidin
Quality Control:
- ACE2 gene expression has been verified by flow cytometry and functional assays.
- TMPRSS2 gene expression has been verified by RT-qPCR and functional assays.
- The expression and induction of ASC::GFP have been verified by Western blot and fluorescence microscopy.
- NLRP1 expression has been verified by Western blot and fluorescence microscopy.
- The stability for 20 passages, following thawing, has been verified.
- These cells are guaranteed mycoplasma-free.
Contents
Please note: Each cell line is sold separately. See TDS for the exact contents of each cell line.
- 3-7 x 106 A549-ASCoV2 cells OR A549-ASCoV2-NLRP1 cells, in a cryovial or shipping flask
- 1 ml of Blasticidin (10 mg/ml)
- 1 ml of Normocin™ (50 mg/ml)
![]() Shipped on dry ice (Europe, USA, Canada and some regions in Asia)
Shipped on dry ice (Europe, USA, Canada and some regions in Asia)
Details
A549 cells for COVID-19 studies
A549 cells are commonly used for the study of respiratory infections. SARS-CoV-2, the causative agent of coronavirus disease-19 (COVID-19), gains entry into the human lung epithelium through the interaction of the virus Spike protein with the host ACE2 and TMPRSS2 receptors [1-3]. A549 cells express negligible levels of ACE2 and no TMPRSS2 and thus are poorly permissive to infection by SARS-CoV-2 [2-4, in-house data]. Stable transfection of A549 cells with the human ACE2 and TMPRSS2 genes increases their permissivity SARS-CoV-2 (in-house data).
A549 cells for inflammasome studies
A549 cells endogenously express proteins involved in the inflammasome signaling including ASC, caspase-1, and Gasdermin D/E. However, they are unable to mount inflammasome responses because of the lack of expression of some sensors/co-sensors. Notably, they lack expression of NLRP1 and NLRP3, two sensors that are strictly dependent on the ASC adaptor to bridge their interaction with pro-caspase-1 (in-house data) [5-7].
The ASC adaptor
ASC (apoptosis-associated speck-like protein containing a CARD domain, aka PYCARD is a protein adaptor important in canonical inflammasome responses [8,9]. ASC's bipartite composition, consisting of one PYD (pyrin domain) and one CARD (caspase recruitment domain) domain, allows the recruitment of the CARD-containing pro-caspase-1 to canonical inflammasome sensors [8]. In resting cells, ASC is present in a soluble and diffuse form, both in the cytoplasm and nucleus. Upon inflammasome activation, ASC molecules form a single large, micrometer-sized, ‘speck’ per cell, thus concentrating caspase-1 activation sites [9].
The NLRP1 sensor
NLRP1 (aka NALP1) was the first described inflammasome sensor of the Nod-Like Receptors (NLR) family [6-9]. Among other NLR, NLRP1 exhibit a unique structure with a PYRIN, NACHT and LRR in its N-terminal domain followed by a FIIND (function-to-find domain) and a CARD domain in its C-terminal region. The FIIND domain autocleaves NLRP1 into two proteins that persist connected in a non-covalent manner and in close association with the dipeptidyl peptidases DPP-8/-9, thereby maintaining NLRP1 in an auto-inhibitory state [6,10]. It took two decades to uncover the activation mechanisms of human NLRP1. The actual model relies on a “functional degradation” pathway [10], in which danger signals trigger proteasomal degradation of the NLRP1 N-terminal domain leading to the release of the C-terminal CARD containing domain, the active form of NLRP1. Subsequently, the ASC adaptor is recruited inducing the assembly of the NLRP1 inflammasome through oligomerization with pro-capase-1 [6-9].
So far, four activators of the NLRP1 pathway have been reported [5, 11-19]:
The firstly identified pathway relies on pharmacological inhibition of the cellular proteases DPP-8/-9 by Val‐boroPro. While the small molecule Val‐boroPro (Talabostat) was known since 2003 to stimulate anti-cancer immune responses [19], only recent work has identified its mechanism of action through activation of the NLRP1 inflammasome [11-13]. However, the physiological settings that trigger NLRP1 activation through DPP-8/DPP-9 inhibition are currently unknown.
The second pathway suggests that NLRP1 is acting as a decoy receptor aiming to detect viral proteases activities. Indeed, enteroviral 3C proteases [14-15] and coronaviral 3CL proteases [5], which are essential for maturation of viral proteins, have been shown to cleave NLRP1 within its N-terminal domain. Cleavage creates an unstable neo-N terminus that is rapidly degraded by the proteasome pathway, leading to NLRP1 inflammasome activation. In the context of SARS-CoV-2 infection, NLRP1 activation by the viral 3CL protease (NSP5) has been linked to lung epithelial cell death and inflammatory cytokine release, suggesting an important role of NLRP1 in COVID-19 pathophysiology [5].
Recent work has identified a third pathway by demonstrating that NLRP1 senses long dsRNA generated in the context of viral infection by Semliki Forrest Virus (SFV) [16]. Interestingly, this pathway can be mimicked by transfection of high molecular weight poly (I:C). It was found that long dsRNAs (>500 bp) bind directly to NLRP1 (NACHT-LRR domain). This interaction activates the ATPase, NACHT domain of NLRP1 leading to its activation following proteasome-dependent degradation of the N-terminal part of NLRP1 [16].
Lastly, Zhong’s team have discovered that human NLRP1 also senses the ultraviolet B (UVB)- and toxin-induced ribotoxic stress response (RSR). Mechanistically, RSR leads to the direct phosphorylation of NLRP1 N-terminal region by the MAP3K ZAK-α and the MAPK p38 hence driving NLRP1 destabilisation and inflammasome activation [17].
Additionally, NLRP1 polymorphism within human population is linked to several rare genetic diseases and autoimmune diseases such as psoriasis, vitiligo or type-1 diabetes [5-6, 20] and thus is considered an important therapeutic target.
1. Wu J, et al., 2021. SARS-CoV-2 ORF9b inhibits RIG-I MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Reports. 34(7):108761.
2. Chen H. et al., 2020. SARS-CoV-2 activated lung epithelial cell proinflammatory signaling and leads to immune dysregulation in COVID-19 patients by single-cell sequencing. medRxiv: DOI 10.1101/2020.05.08.20096024.
3. Hoffmann M. et al., 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:1-16.
4. Matsuyama S. et al., 2020. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. PNAS. 117(13):7001-7003.
5. Planès R. et al., 2022. Human NLRP1 is a sensor of pathogenic coronavirus 3CL proteases in lung epithelial cells. Mol Cell. S1097-2765(22).
6. Taabazuing C.Y. et al., 2020. NLRP1 ha The NLRP1 and CARD8 inflammasomes. Immunol. Reviews. 297(1):13-25.
7. Swanson K.V. et al., 2019. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19:477-489.
8. Mathur A. et al., 2017. Molecular mechanisms of inflammasome signaling. J. Leuk. Biol. 103:233.
9. Hoss F. et al., 2017. Assembly and regulation of ASC specks. Cell. Mol. Life Sci. 74:1211.
10. Sandstrom et al., 2019. Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science. 2019 Apr 5;364(6435):eaau1330. doi: 10.1126/science.aau1330. Epub 2019 Mar 14. PMID: 30872533; PMCID: PMC6532986.
11. Okondo et al., 2017. DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat Chem Biol. 2017 Jan;13(1):46-53. doi: 10.1038/nchembio.2229. Epub 2016 Nov 7. PMID: 27820798; PMCID: PMC5477230.
12. Taabazuing et al., 2017. Pyroptosis and Apoptosis Pathways Engage in Bidirectional Crosstalk in Monocytes and Macrophages. Cell Chem Biol. 2017 Apr 20;24(4):507-514.e4. doi: 10.1016/j.chembiol.2017.03.009. Epub 2017 Apr 6. PMID: 28392147; PMCID: PMC5467448.
13. Zhong et al., 2018. Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J Biol Chem. 2018 Dec 7;293(49):18864-18878. doi: 10.1074/jbc.RA118.004350. Epub 2018 Oct 5. PMID: 30291141; PMCID: PMC6295727.Robinson et al., 2022.
14. Tsu et al., 2021. Diverse viral proteases activate the NLRP1 inflammasome. Elife. 2021 Jan 7;10:e60609. doi: 10.7554/eLife.60609. PMID: 33410748; PMCID: PMC7857732.
15. Robinson et al., 2020. Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science. 2020 Dec 4;370(6521):eaay2002. doi: 10.1126/science.aay2002. Epub 2020 Oct 22. PMID: 33093214.
16. Bauernfried et al., 2022. Human NLRP1 is a sensor for double-stranded RNA. Science. 2021 Jan 29;371(6528):eabd0811. doi: 10.1126/science.abd0811. Epub 2020 Nov 26. PMID: 33243852.
17. Robinson et al., 2022. ZAKα-driven ribotoxic stress response activates the human NLRP1 inflammasome. Science. 2022 Jul 15;377(6603):328-335. doi: 10.1126/science.abl6324. Epub 2022 Jul 14. PMID: 35857590.
18. Walsh et al., 2003. Val-boroPro accelerates T cell priming via modulation of dendritic cell trafficking resulting in complete regression of established murine tumors. PLoS One. 2013;8(3):e58860. doi: 10.1371/journal.pone.0058860. Epub 2013 Mar 12. PMID: 23554941; PMCID: PMC3595211.
19. Adams et al., 2004. PT-100, a small molecule dipeptidyl peptidase inhibitor, has potent antitumor effects and augments antibody-mediated cytotoxicity via a novel immune mechanism. Cancer Res. 2004 Aug 1;64(15):5471-80. doi: 10.1158/0008-5472.CAN-04-0447. PMID: 15289357.
20. Levandowski et al., 2013. NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1β processing via the NLRP1 inflammasome. Proc Natl Acad Sci U S A. 2013 Feb 19;110(8):2952-6. doi: 10.1073/pnas.1222808110. Epub 2013 Feb 4. PMID: 23382179; PMCID: PMC3581876.